
Direct identification of T cell epitopes in cancer tissues
Pancreatic ductal adenocarcinoma (PDAC), the most common type of pancreatic cancer, is one of the most lethal cancers, and its 5-year survival rate is 12% (1). The lack of reliable early diagnostic markers and the rapid development of resistance to conventional therapies are among the challenges to the management of this malignant disease (2).
Prediction of cancer-specific T cell epitopes is an important part of cancer immunotherapies. Discovering new potential immunotherapy targets is crucial for improving PDAC treatment outcomes to immunotherapy. T cells are the major effector cells for antitumor adaptive immune responses. T cells recognize tumor cells by using the T cell receptor (TCR) to bind to antigenic epitopes presented by tumor cells or through antigen cross-presentation by antigen-presenting cells. This binding is highly specific and is also restricted by the human leukocyte antigens (HLA) on tumor cells or antigen-presenting cells (3). It has been a challenge to identify T cell epitopes on pancreatic cancer antigens, largely due to the lack of knowledge on immunodominant antigens in PDAC (4) and the lack of effective technical approaches.
However, T cell epitopes have only been characterized in a few of such antigens including NY-ESO-1 (5), MAGE-A (6,7), and Wilms’ tumor gene 1 (WT1) (8,9); and TCR-T-cell therapies targeting these antigens are still under clinical evaluation. Alternatively, T cell epitopes were identified on the antigens by analyzing the T cells induced by the administration of cancer vaccines that are comprised of the antigens (10,11). Such an approach would limit the T cell epitope discovery in a small number of patients who were treated with experimental cancer vaccines.
More recently, after recognizing the value of mutation-associated neoepitopes, in silico epitope prediction from the whole-exome sequencing (WES) results has become another approach (12,13). However, this approach may not be able to predict any high-affinity TCR binding epitopes if the tumors have low tumor mutation burdens (TMB) (14). In addition, the binding of the predicted epitopes to HLA molecules and TCR would still need to be validated (15).
Mass spectrometry (MS) was used to identify the T cell epitopes by eluting the peptides from HLA molecules. Kalaora et al. performed both WES and direct-peptide identification with MS and selected overlapping peptide sequences predicted by both DNA mutations and identified by MS (16). This approach of identifying neoepitopes was demonstrated to be feasible with melanoma tissue specimens (17). However, this approach has not been tested in tumors with low TMB, such as PDAC (18). It was recently shown that genomic mutation-derived HLA-class I epitopes were rarely presented in hepatocellular carcinomas, which have a relatively low TMB as compared to other malignancies with high TMB, such as malignant melanoma (14). Fujiwara et al. are the first ones to use MS to identify HLA class I and class II-restricted peptidomes in human PDAC (Figure 1). Briefly, the protein lysate from PDAC specimens and cell lines were subjected to the antibody affinity purification of human major histocompatibility complexes (MHC) including both HLA class I and class II complexes (Figure 2). Peptides bound to the MHC were then eluted and identified through LC-MS/MS. A few novel findings were made with this study. T cells that recognize shared antigens exist in the circulating system of PDAC patients and are also likely in the tumors. This study has identified the peptide sequences that are HLA class I epitopes shared by PDACs from patients with different HLA types and those that are overlapped between HLA class I and HLA class II epitopes, respectively (Table 1). Nevertheless, tetramer staining is warranted to further validate in vivo the presence of epitope-specific T cells once the epitope-specific tetramers become available. Some of these peptides that are from potential anti-cancer targets were selected for further validation for their binding to class I HLA molecules and their ability in inducing cytokine expression in T cells. The peptides, particularly those that can induce polyfunctional T cells, may be therapeutic targets for cancer vaccine and T cell therapy.
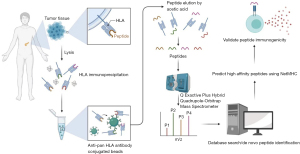
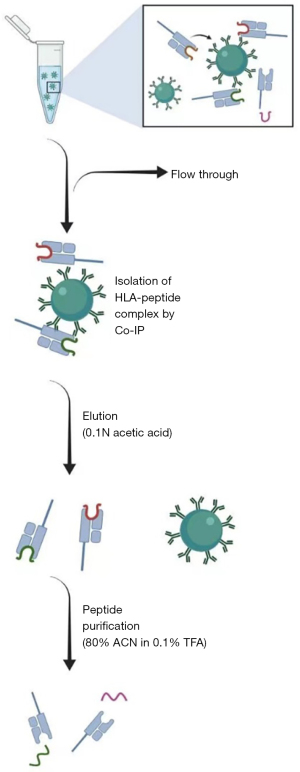
Table 1
| Epitopes shared among multiple HLA types |
| COL6A3, ELOVL1, LAMC2, RASAL2, DYNLRB1, ICE1, LAMB3, MYH9, ORMDL3, MYL12A, WDR82, TRRAP, TFIP11, ACBD3, CKS2, IGF1, TRAPPC11, ZMYND11, CTNNBIP1 |
| Epitopes shared by HLA class I and class II types |
| FGA, IGHG, TMSB10, VIM, HBD, HIF2 |
HLA, human leukocyte antigens.
Interestingly, this study has not identified any epitope that corresponds to the genomic variants. They cannot exclude the possibility that genomic variant-associated neoepitopes exist rarely and thus were missed. However, Fujiwara et al. did identify peptide variants. How these peptide variants have been generated remains to be investigated in the follow-up studies. The peptidome datasets generated in this study may help improve the algorithm of the in-silico prediction of HLA class I and HLA class II epitopes.
Fujiwara et al. found that identified epitopes can bind unmatched HLA molecules and induce T cell response in peripheral T cells from HLA-type unmatched patients. Although the exact mechanism remains to be investigated, it is conceivable that there are homologies among different HLA class I molecules and even possibly among HLA-type unmatched TCRs. More importantly, Fujiwara’s results suggest that it is possible to develop cancer vaccines and TCR T cell therapies with the epitopes identified for all patients regardless of their HLA types. Such a notion is being validated with a follow-up study of testing the in vivo antitumor activities of TCRs that recognize such shared epitopes. HLA class II epitopes were not well studied in the past (19,20). The study by Fujiwara et al. may provide the HLA class II peptidome dataset for further investigation on how to predict the binding between HLA class II molecules and epitopes. This study is limited by having not validated the binding strength between identified HLA class II epitopes and HLA class II molecules. Therefore, when Fujiwara et al. narrowed down the epitopes for T cell stimulation assays, they chose those that were predicted to have the best binding affinities with HLA class II molecules. Considering that MS-identified epitopes are specifically presented in vivo by the HLA class II MHC complex, the current algorithm such as NetMHC and NetMHCIIpan may have missed the best HLA class II binding epitopes. Nevertheless, they demonstrated that the peptides that contain overlapped HLA class I and HLA class II epitopes can induce the response in polyfunctional T cells. Therefore, this approach will possibly identify the HLA class II epitopes appropriate for cancer vaccines and T cell therapies.
A few other limitations in the study by Fujiwara et al. should be recognized. First, the study was not able to distinguish between the epitopes from tumor epithelial cells or stromal cells. We did the epitope discovery with PDAC tumor cell lines and were able to identify a large number of epitopes overlapping with those identified in human PDAC tissues. They need to compare the epitopes identified in tumor tissues to those identified in paratumoral normal tissues in PDACs similarly to a previously published study (21). Second, identifying epitopes by MS is technically tedious. Whether this approach is reproducible among different laboratories and different investigators remains to be tested. Recently, Jaeger et al. used genetically engineered mouse models for identifying PDAC tumor-specific MHC class I peptides (22), thus, providing a venue for studying the immunopeptidome reproducibly in the mouse model of PDAC. Fujiwara et al. did not observe any epitopes from known PDAC-associated antigens (23,24). It is possible that epitopes from known PDAC-associated antigens are tolerogenic and also possible that Fujiwara’s approach has missed those epitopes technically. Among eluted HLA class I bound peptides, they have focused on studying 9-mer peptides. However, 10 and 11-mer peptides, which are predicted to bind HLA class I albeit at a lesser strength, warrant future studies.
In summary, studies on the direct identification of T cell epitopes will facilitate a further understanding of the mechanisms of the binding between MHC complexes, TCR, and epitopes. More importantly, such studies have led to ongoing effort in the preclinical development of off-shelf productions of cancer vaccine and T cell therapy for the shared epitopes identified here.
Acknowledgments
Funding: This work was supported by Sidney Kimmel Comprehensive Cancer Center Grant NIH P30 CA006973, and was supported in part by a JSPS Overseas Research Fellowship from the Japan Society for the Promotion of Science (to K Fujiwara).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://apc.amegroups.com/article/view/10.21037/apc-2023-1/coif). KF was supported by a JSPS Overseas Research Fellowship from the Japan Society for the Promotion of Science. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17-48. [Crossref] [PubMed]
- Coleman O, Henry M, O'Neill F, et al. A Comparative Quantitative LC-MS/MS Profiling Analysis of Human Pancreatic Adenocarcinoma, Adjacent-Normal Tissue, and Patient-Derived Tumour Xenografts. Proteomes 2018;6:45. [Crossref] [PubMed]
- Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1-10. [Crossref] [PubMed]
- Yadav M, Jhunjhunwala S, Phung QT, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014;515:572-6. [Crossref] [PubMed]
- Hunder NN, Wallen H, Cao J, et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med 2008;358:2698-703. [Crossref] [PubMed]
- van der Bruggen P, Traversari C, Chomez P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 1991;254:1643-7. [Crossref] [PubMed]
- Zajac P, Schultz-Thater E, Tornillo L, et al. MAGE-A Antigens and Cancer Immunotherapy. Front Med (Lausanne) 2017;4:18. [Crossref] [PubMed]
- Cheever MA, Allison JP, Ferris AS, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res 2009;15:5323-37. [Crossref] [PubMed]
- van Amerongen RA, Hagedoorn RS, Remst DFG, et al. WT1-specific TCRs directed against newly identified peptides install antitumor reactivity against acute myeloid leukemia and ovarian carcinoma. J Immunother Cancer 2022;10:e004409. [Crossref] [PubMed]
- Lutz E, Yeo CJ, Lillemoe KD, et al. A lethally irradiated allogeneic granulocyte-macrophage colony stimulating factor-secreting tumor vaccine for pancreatic adenocarcinoma. A Phase II trial of safety, efficacy, and immune activation. Ann Surg 2011;253:328-35. [Crossref] [PubMed]
- Thomas AM, Santarsiero LM, Lutz ER, et al. Mesothelin-specific CD8(+) T cell responses provide evidence of in vivo cross-priming by antigen-presenting cells in vaccinated pancreatic cancer patients. J Exp Med 2004;200:297-306. [Crossref] [PubMed]
- Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017;547:222-6. [Crossref] [PubMed]
- Balachandran VP, Łuksza M, Zhao JN, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017;551:512-6. [Crossref] [PubMed]
- Löffler MW, Mohr C, Bichmann L, et al. Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma. Genome Med 2019;11:28. [Crossref] [PubMed]
- Hopkins A, Jaffee E. Pancreatic cancer: Next-generation algorithms for neoantigen selection. Nat Rev Gastroenterol Hepatol 2018;15:135-6. [Crossref] [PubMed]
- Kalaora S, Barnea E, Merhavi-Shoham E, et al. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget 2016;7:5110-7. [Crossref] [PubMed]
- Bassani-Sternberg M, Bräunlein E, Klar R, et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 2016;7:13404. [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Abelin JG, Harjanto D, Malloy M, et al. Defining HLA-II Ligand Processing and Binding Rules with Mass Spectrometry Enhances Cancer Epitope Prediction. Immunity 2019;51:766-779.e17. [Crossref] [PubMed]
- Racle J, Michaux J, Rockinger GA, et al. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat Biotechnol 2019;37:1283-6. [Crossref] [PubMed]
- Schuster H, Peper JK, Bösmüller HC, et al. The immunopeptidomic landscape of ovarian carcinomas. Proc Natl Acad Sci U S A 2017;114:E9942-51. [Crossref] [PubMed]
- Jaeger AM, Stopfer LE, Ahn R, et al. Deciphering the immunopeptidome in vivo reveals new tumour antigens. Nature 2022;607:149-55. [Crossref] [PubMed]
- Hassan R, Thomas A, Alewine C, et al. Mesothelin Immunotherapy for Cancer: Ready for Prime Time? J Clin Oncol 2016;34:4171-9. [Crossref] [PubMed]
- Akce M, Zaidi MY, Waller EK, et al. The Potential of CAR T Cell Therapy in Pancreatic Cancer. Front Immunol 2018;9:2166. [Crossref] [PubMed]
Cite this article as: Shao Y, Zhang T, Celiker B, Fujiwara K. Direct identification of T cell epitopes in cancer tissues. Ann Pancreat Cancer 2023;6:3.


