
Genetic alterations in the neuronal development genes are associated with changes of the tumor immune microenvironment in pancreatic cancer
Highlight box
Key findings
• The role of neuronal development genes in the tumor immune microenvironment (TME) of pancreatic cancer.
What is known and what is new?
• Neuronal development genes are associated with perineural invasion and poor prognosis of pancreatic ductal adenocarcinoma (PDAC).
• Alterations of neuronal development genes are associated with changes in the densities of major immune cells in TME of PDAC.
What is the implication, and what should change now?
• It remains to be further investigated how neuronal development genes modulate the TME of PDAC.
Introduction
The annual incidence of pancreatic cancer is increasing worldwide and is projected to become the second-leading cause of cancer death by 2040 in the US (1). Pancreatic ductal adenocarcinoma (PDAC), accounting for approximately 90% of pancreatic cancer, is an aggressive disease characterized by a dismal outcome with a 5-year survival rate of 12% (2). The poor prognosis can be attributed to delayed diagnosis, invasive tumor nature, frequent metastasis, and high resistance against all conventional therapies (3). The tumor immune microenvironment (TME) surrounding PDAC cells significantly determines tumor growth, metastatic ability, and treatment resistance (4). Additionally, accumulating evidence indicates the important role of neural signaling, neural regulation, and neurotransmitters in the TME and development of PDAC (5-7). In 2012, the International Cancer Genomics Consortium (ICGC) found that PDAC was enriched with axon guidance gene family genetic alterations. This suggests the potential involvement of the nervous system in PDAC carcinogenesis and has led to a rising interest in this aspect of neuronal mechanism (8).
Axon guidance family molecules have been studied for their roles in angiogenesis, tumorigenesis, and immune regulation (9-12). Semaphorins (SEMA) are a large family of axon guidance molecules and have been recognized as critical contributors to neural development, the immune response, and tumor progression (13). In our previous studies, PDAC cells secreted Sema3D, which is regulated by Annexin A2 (ANXA2) and binds Plexin D1 (PLXND1) and neuropilin-1 through an autocrine signaling mechanism. This binding interaction was also shown to promote invasion and metastasis of PDAC (14,15). In addition, we also proved that the SEMA3D-PLXND1 axon guidance pathway mediates paracrine signaling between tumor cells and nerves to enhance innervation and perineural invasion (PNI) in PDAC (16). Our group found that nerve-derived Sema3D and potentially other SEMA promote tumor progression and metastasis by reprograming tumor-associated macrophages toward M2 polarization. In a recent single-nucleus and spatial transcriptome profiling study of PDAC (17), 20 neuronal development genes [disks large homolog 2 (DLG2), neuron-glial-related cell adhesion molecule (NRCAM), neurexin3 (NRXN3), mitogen-activated protein kinase 10 (MAPK10), platelet-derived growth factor D (PDGFD), PRKCE, potassium calcium-activated channel subfamily M alpha 1 (KCNMA1), polycystic kidney and hepatic disease 1 (PKHD1), neural cell adhesion molecule 1 (NCAM1), neuregulin-1 (NRG1), zinc finger protein 667 (ZNF667), cystic fibrosis transmembrane conductance regulator (CFTR), acyl-CoA medium-chain synthetase-3 (ACSM3), complement 6 (C6), protein tyrosine phosphatase receptor type M (PTPRM), hypoxia-inducible factor 1 alpha (HIF1A), adenylyl cyclase 5 (ADCY5), adherens junctions-associated protein 1 (AJAP1), neurobeachin (NBEA), sodium voltage-gated channel alpha subunit 9 (SCN9A)] were found to be associated with PNI and poor prognosis in their independent cohorts. We aim to further investigate the findings of Hwang et al. (17) by examining the relationships between specific genetic alterations in neuronal development genes and the TME.
Methods
Patient specimens
Specimens from 89 pancreatic cancer patients treated at the Johns Hopkins Hospital, comprised of 41 surgically resectable and 48 locally advanced pancreatic cancer (LAPC) patients, were included in the study (available at https://cdn.amegroups.cn/static/public/10.21037apc-23-13-1.xlsx). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of Johns Hopkins Medicine (J1568). These specimens were archived from two clinical trials (NCT02451982; NCT02648282), and written informed consent to allow the usage of archived specimens for other studies (the Johns Hopkins Medical Institution Institutional Review Board approved protocols at the Johns Hopkins Pancreatic Cancer Precision Medicine Center of Excellence Program) including this study was obtained from all the patients. Specimens were either biopsy specimens collected via endoscopic ultrasound-guided fine needle core biopsies or surgically resected tumors. The demographics of the human cohort were described in the studies by Li et al. (18).
Sequential multiplex immunohistochemistry (mIHC)
The sequential mIHC was performed on biopsy specimens before the patients received any experimental therapy, the method to analyze the percentage of immune cell infiltration, and markers to identify immune cells, as described in the study by Li et al. (18). The published quantitative results were used without any modification for the analysis in this study. Sixty-three out of 89 patients have mIHC quantitative results available for this study.
Statistical analysis
Welch’s t-test was performed to compare the two groups. We assume the sample means being compared for two normally distributed populations, and the populations have equal variances. A P value <0.05 was considered statistically significant. Statistical analyses and graphs were generated using GraphPad Prism v.9.5.1.
Results
A decrease in intratumoral CD8+ T cells is associated with the genetic alteration of neuronal development genes
Following our previous studies suggesting axon guidance molecules such as the class 3 SEMA modulate the TME, we attempted to understand how the genetic alteration of axon guidance genes correlates with the infiltration of immune cell subtypes in PDACs. In addition to those axon guidance genes reported to be associated with PDACs by our group and others, Hwang et al. (17) recently identified 20 PDAC-associated neuronal development genes through a comprehensive approach. We, therefore, analyzed the genetic alterations in these 20 neuronal development genes in our published PDAC patient cohorts whose intratumoral immune infiltrates have been characterized. The whole exome sequencing (WES) and whole genomic sequencing (WGS) data were obtained from the Precision Medicine Application Platform at Johns Hopkins, where the WES and WGS data are deposited. Due to the lack of functional assays to determine the significance and functions of the missense mutations, we focused on the copy number gain or loss in this study. Fifty-two of 89 PDACs have both WGS and WES data; for these cases, we determined the copy number gain (3 or more copies) or loss (0 or 1 copy) by using the WGS data (available at https://cdn.amegroups.cn/static/public/10.21037apc-23-13-1.xlsx). The remaining 37 PDACs have only WES data; we determined the copy number gain or loss using the WES data for these cases. We performed Welch’s t-test to compare the subgroup with the copy number gain or loss with the subset without either gain or loss (Tables S1,S2).
In many of these 20 genes, either there is a gain or loss, and CD8+ T cells were significantly decreased compared to PDAC without gain or loss of these genes (Figure 1). Nevertheless, the loss was more likely and significantly associated with decreased intratumor CD8+ T cells. The only exception is that loss of ZNF667 is associated with an increase in CD8+ T cells.
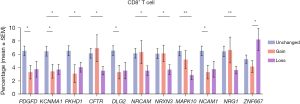
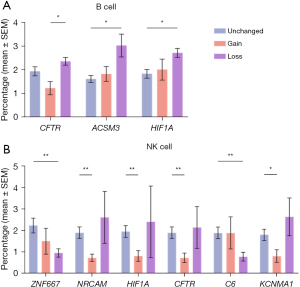
An increase in intratumoral B cells and a decrease in intratumoral natural killer (NK) cells is associated with the loss of neuronal development genes
An increase in intratumoral B cells is associated with the loss of CFTR, ACSM3, and HIF1A. In contrast, gain in any of these 20 genes is not associated with a significant change in B cell infiltration (Figure 2A). The only exception is that CFTR gain is associated with decreased B cell infiltration. NK cells vary in association with axon guidance genes (Figure 2B). However, a significant reduction of NK cells is associated with the gain of NRCAM, KCNMA1, HIF1A, and CFTR genes. In contrast, a significant decrease in NK cells is associated with the loss of ZNF667 and C6.
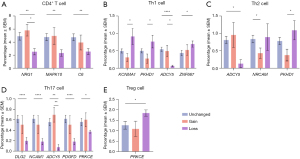
Genetic alterations in neuronal development genes are correlated with changes in CD4+ T cell infiltration, including their subtypes
A significant decrease in intratumoral CD4+ T cell infiltration is associated with the loss of the MAPK10, NRG1, and C6 genes (Figure 3). In contrast, a significant increase in the T helper 1 (Th1) subtype is associated with the loss of the ZNF667, KCNMA1, and PKHD1 gene, and a significant decrease in the Th2 subtype is associated with the gain of the NRCAM and PKHD1 genes. A significant decrease in the Th17 subtype is associated with the loss of DLG2, PDGFD, protein kinase C epsilon (PRKCE), ADCY5 and NCAM1. The only significant increase in the regulatory T (Treg) subtype is associated with the loss of PRKCE. Interestingly, a considerable decrease in Th1, Th2, and Th17 infiltrations is related to the loss of ADCY15.
Genetic alterations in neuronal development genes are associated with T cell exhaustion
A significant decrease in the PD-1+CD4+ T cells is associated with the loss of DLG2, PDGFD, ADCY15, and NCAM1, while a significant decrease in the PD-1+CD8+ T cells is associated with the loss of ADCY15 and MAPK10 (Figure 4). A significant decrease in the EOMES+CD8+ T cells is associated with the loss of NRXN3.

A significant decrease in M1-like tumor-associated macrophages (TAM), M2-like TAM, and tumor-associated neutrophils (TAN) is associated with the loss of neuronal development genes
A significant decrease in M1-like TAM infiltration is associated with the loss of C6, ADCY5, and SCN9A. A significant decrease in the infiltration of M2-like TAM is associated with the loss of NRXN3, C6, HIF1A, ADCY5, and SCN9A (Figure 5). Thus, a decrease in both M1-like and M2-like TAMs is associated with the loss of the same genes. It should be noted that a significant reduction in the CD66b+ TAN is associated with the loss of ZNF667.

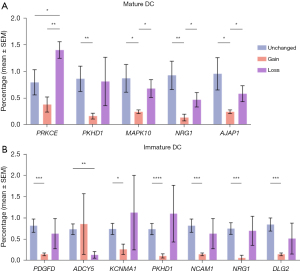
Changes in dendritic cell (DC) count are linked to genetic alterations in neuronal development genes
A significant decrease in the immature DCs is associated with the gain of DLG2, PDGFD, NCAM1, NRG1, NCAM1, and PKHD1 genes, and a significant decrease in the mature DCs is associated with the gain of MAPK10, PRKCE, PKHD1, NRG1, and AJAP1 genes (Figure 6). It should be noted that a significant decrease in the immature DCs is associated with the loss of ADCY5 and that a significant increase in the mature DCs is associated with the loss of PRKCE.
Discussion
In this study, we performed a comprehensive analysis of the association of the PDAC-related neuronal development genes with the modulation of major immune cell subtypes in the tumor microenvironment of PDAC. Except for PTPRM and NBEA, genetic alterations involving this 20-gene panel are associated with significant changes in specific immune cell subtypes. Except for AJAP1, the loss involving this panel of neuronal development genes is significantly associated with changes in immune cells. In contrast, gain in specific genes, including NRXN3, ZNF667, ACSM3 C6, ADCY5, SCN9A, and PRKCE, is significantly associated with changes in immune cells. Where there is a loss involving this panel of neuronal development genes, a decrease was noted in most immune cell subtypes examined. Among the immune cell subtypes, CD8+ T cells were most commonly decreased due to the loss in the neuronal development genes, followed by Th17, M2-like TAM, and PD-1+CD4+ T cells. Interestingly, where there is a gain in these neuronal development genes, only a decrease was observed as a statistically significant change in the immune infiltrates across all the immune cell subtypes examined. Additionally, immature DCs are the most commonly decreased subtype, followed by mature DC, CD8+ T cells, and NK cells. Our results suggest that genetic alterations in the neuronal development genes in PDAC lead to a decrease in adaptive and effector cell immune response. Although some of these neuronal development genes were implicated in the immune modulation in literature as described below- for most of these genes, it is the first time that their potential roles in the TME of PDACs have been demonstrated.
The DLG2 gene encodes an excitatory postsynaptic scaffold protein, which is a member of the membrane-associated guanylate kinase (MAGUK) family and is abundantly expressed in brain tissues (19). In the transcriptomic profiling studies, researchers found the expression of DLG2 in mast cells, splenic red pulp macrophages, and plasmacytoid DCs that produce interferons-β (20). However, there was a lack of data supporting the role of the DLG2 expression in the TME, except for one study showing that DLG2 stimulated inflammasome formation and increased apoptosis in macrophage-like cells (21). Our results show that loss of DLG2 was significantly associated with a decrease of intratumoral PD-1+CD4+ T cells, Th17, and CD8+ T cells in PDAC, and gain of DLG2 was significantly associated with an increase of immature DCs and CD8+ T cells.
The NRCAM gene encodes a transmembrane protein composed of multiple immunoglobulin-like C2 domains and a fibronectin type III domain. NRCAM plays an essential role in neuronal development, including neuron-neuron adhesion, axon growth, signal transduction, synapse formation, and formation of myelinated nerve structures (22,23). Only one study reported that the differentiation of Th1 cells was affected by regulating NRCAM gene transcription in Graves’ disease (24). Our results showed that loss of NRCAM was significantly associated with decreased CD8+ T cells in PDAC; gain of NRCAM was significantly associated with reduced NK cells and Th2 cells.
NRXN3 gene, a member of the neurexin gene family, is a synaptic modulator and encodes a protein that functions in the vertebrate nervous system as a cell adhesion molecule during synaptogenesis and intercellular signaling (25-27). The role of NRXN3 in the TME has not been previously studied. Our results show that loss of NRXN3 was significantly associated with a decrease of CD8+ T cells, EOMES+CD8+ T cells, and M2-like TAMs in PDAC, and gain of NRXN3 was significantly associated with an increase of M2-like TAMs.
MAPK10 involves various processes such as neuronal proliferation, differentiation, migration, and programmed cell death. In a recently published study of ovarian cancer, expression of MAPK10 has been found to be positively correlated with the infiltration of eosinophils and naive B cells and negatively correlated with the infiltration of NK cells and memory B cells (28). Another study of breast cancer showed that mutations in MAPK10 were associated with a decreased infiltration of activated CD4+ T cells (29). Consistently, our results showed that loss of MAPK10 was significantly associated with a decrease of CD4+ T cells, PD-1+CD8+ T cells, and mature DCs in PDAC; gain of MAPK10 was significantly associated with an increase of PD-1+CD8+ T cells and mature DCs.
NCAM1 is a synaptic adhesion molecule that supports synaptic connections by trans-homophilic binding. NCAM1 has been implicated in several brain-related biological processes, including neuronal migration, axonal branching, fasciculation, and synaptogenesis, with a pivotal role in synaptic plasticity. In human hematopoiesis, NCAM1 (also called CD56) is highly expressed in NK cells (30,31). In addition, NCAM1 expression was found in rare subsets of T and B lymphocytes, DCs, and neural or mesenchymal stem cells (32). However, no study evaluates the immune-modulating effect of NCAM1. Our results showed that loss of NCAM1 was significantly associated with a decrease of Th17 and PD-1+CD4+ T cells in PDAC; gain of NCAM1 was significantly associated with an increase of CD8+ T cells and immature DCs.
NRG1 belongs to the epidermal growth factor (EGF) family, contains an EGF-like domain, and is a HER3 (ERBB3) ligand. NRG1 is primarily known for its essential role in Schwann cell and oligodendrocyte differentiation, maintenance, and myelination in the central and peripheral nervous systems (33). Furthermore, through the NRG1-HER3 signaling axis, NRG1 contributes to malignant tumor development in several cancer types, including gastric, pancreatic, breast cancer, squamous cell carcinoma, and non-small-cell lung cancer. Additionally, its overexpression is closely associated with poor prognosis (34-36). A recent study indicated that NRG1 augments regulatory populations of macrophages, T cells, and B cells peripherally and in injured spinal cord tissue (37). In a study of colorectal cancer, NRG1 expression was positively correlated with activated DCs, neutrophils, plasma cells, and resting CD4+ memory T cells and negatively with memory B cells and macrophage M1 (38). In addition, a study of oral squamous cell carcinoma also showed that NRG1 expression was negatively correlated with the infiltration of B cells and CD8+ T cells (39). Our results show that loss of the NRG1 gene was significantly associated with a decrease of CD4+ T cells in PDAC; gain of NRG1 was significantly associated with a decrease of mature DCs and immature DCs.
ZNF667 is a novel C2H2 zinc finger protein that is found to be significantly upregulated during myocardial and cerebral ischemic preconditioning (40,41). A recent study showed that ZNF667 exhibited anti-inflammatory effects in LPS-induced macrophages by suppressing the mTOR-dependent expression of glycolytic genes and glycolysis (42). Our results show that loss of ZNF667 is significantly associated with a decrease of NK cells and TAN and an increase of CD8+ T cells in PDAC.
HIF1A is involved in cellular metabolism, cell death, and proliferation of neuronal progenitors in the sympathetic system (43,44). HIF1A is a crucial regulator of immune cell function in health and disease (45). HIF1A regulates M1 macrophage polarization, DC maturation and migration, and neutrophil NET formation and survival (46,47). Our results showed that loss of HIF1A was significantly associated with a decrease of M2-like TAMs and an increase of B cells in PDAC. In contrast, a gain of HIF1A was significantly associated with reduced NK cells.
PDGFD gene belongs to the PDGF family of proteins, which can regulate neurogenesis and diverse functions in the brain. A recent study found that exogenous administration of PDGFB, which shares the same binding receptor with PDGFD, promotes the proliferation of neural progenitor cells (48). Another study found that PDGFD significantly correlates with plasma cells, CD4 memory T cells, follicular helper T cells, monocytes, macrophage M0 and M1, DCs, and mast cells in gastric cancer (49). Our results showed that loss of PDGFD was significantly associated with a decrease of Th17 and PD-1+CD4+ T cells in PDAC; gain of PDGFD was significantly associated with a decrease of CD8+ T cells and immature DCs.
ACSM3 is a member of the acyl-CoA synthase gene family, which regulates cellular phospholipid acyl-chain diversity in the brain (50). A study of melanoma found that ACSM3 was positively correlated with central memory and naïve CD8+ cells, regulatory T cells, macrophages, and DCs (51). Our study found that loss of ACSM3 was significantly associated with an increase of B cells in PDAC.
AJAP1 is an integral transmembrane protein that interacts with the E-cadherin-catenin complex at adherens junctions, which mediate adhesion between pre-and postsynaptic membranes (52). The role of the AJAP1 expression in the TME was not previously reported. Our study found that the gain of AJAP1 was significantly associated with a decrease of mature DCs and Th1 cells in PDAC.
KCNMA1 gene encodes a protein that forms large-conductance calcium-activated potassium channels in cells. These channels play a crucial role in regulating the flow of potassium ions across cell membranes, an essential step in various physiological processes, including neurotransmitter release (53). Mutations of the KCNMA1 gene can lead to channel dysfunction and are associated with neurological conditions, including seizures, movement disorders, developmental delay, and intellectual disability (54). The role of KCNMA1 in the TME was not reported. Our results showed that loss of KCNMA1 was significantly associated with a decrease of CD8+ T cells and an increase of Th1 cells in the tumor microenvironment of PDAC; gain of KCNMA1 was significantly associated with a decrease of CD8+ T cells, NK cells, and immature DCs.
PKHD1 gene encodes a protein known as fibrocystin/polyductin, which is primarily found in the primary cilia of renal epithelial cells. Mutations of the PKHD1 gene are associated with autosomal recessive polycystic kidney disease (ARPKD), a rare genetic disorder characterized by the formation of cysts in the kidneys and liver (55,56). During embryogenesis, PKHD1 is widely expressed in epithelial derivatives, including neural tubules (57). The role of PKHD1 in the TME has not been previously studied. Our results found that loss of PKHD1 was significantly associated with a decrease of CD8+ T cells and an increase of Th1 and Th2 cells in PDAC; gain of PKHD1 was significantly associated with a decrease of mature DCs, immature DCs, and Th2 cells.
CFTR gene encodes channel proteins that belong to the ATP-binding cassette transporter superfamily. CFTR binds ATP and promotes substrate transport across membranes. Mutations in the CFTR gene are the cause of cystic fibrosis. CFTR has been reported to be involved in modulating neuronal excitability through chloride transporters in both the peripheral and central nervous systems (58). One study on cystic fibrosis showed that CFTR regulated B cell activation and lymphoid follicle development. However, the role of CFTR in the TME was not studied. Our results showed that loss of CFTR was significantly associated with a decrease of CD8+ T cells in PDAC; gain of CFTR was significantly associated with a decrease of NK cells.
The C6 gene encodes a component of the complement cascade as a part of the membrane attack complex that can be incorporated into the cell membrane and cause cell lysis. C6 is part of the immune system and is crucial in defending the body against infections and pathogens. In addition to being a component of innate and adaptive immunity, complement proteins regulate several physiologic processes, including synaptic pruning during brain development (59-62). However, the role of the C6 expression in the TME was not well studied. Our results showed that loss of C6 was significantly associated with a decrease of NK cells, CD4+ T cells, M1-like TAM, and M2-like TAMs in PDAC.
ADCY5 is a member of membrane-bound adenylyl cyclase enzymes that regulate cellular activities by mediating G protein-coupled receptor signaling through synthesizing the second messenger cAMP (63). ADCY5 is highly expressed in the brain and myocardium (64). Mutations in ADCY5 have mainly been linked to various complex movement disorders often associated with neurodevelopmental phenotypes (65). However, the role of ADCY5 in the TME was not reported. Our results showed that loss of ADCY5 was significantly associated with a decrease of immature DCs, Th1, Th2, Th17, PD-1+CD4+ T cells, PD-1+CD8+ T cells, M1-like TAMs, M2-like TAMs, and immature DCs in PDAC.
SCN9A, known as Nav1.7, are voltage-gated sodium channels that are preferentially expressed in the dorsal root ganglia and sympathetic neurons (66-68). SCN9A mediates cellular excitability and plays a crucial role in gating pain transmission from the periphery to the central nervous system (69,70). The role of SCN9A in the TME was not reported. Our results showed that loss of SCN9A was significantly associated with a decrease of M1-like TAMs and M2-like TAMs in PDAC.
PRKCE belongs to a family of serine- and threonine-specific protein kinases that can be activated by calcium and the second messenger, diacylglycerol. The gene product of PRKCE plays an essential role in regulating multiple cellular processes, including neuron growth and immune response (71,72). Our results show that loss of PRKCE was significantly associated with a decrease of Th17 cells and an increase of Treg cells and mature DCs in PDAC.
PTPRM is a member of the protein tyrosine phosphatase (PTP) family (73). PTPRM is a key regulator of neurite outgrowth and synapse formation in cortical neurons (74,75). NBEA is a kinase anchor protein that contributes to the regulation of protein trafficking and recycling of ionotropic glutamate and GABA receptors (76,77). NBEA has also been implicated in vesicular traffic at the synapse and has been shown to be required for normal development of the synapses (78). The role of PTPRM or NBEA in the TME was not reported. Our results showed that neither the genetic alterations of PTPRM nor NBEA were significantly associated with a change in immune cells in PDAC.
The limitation of this study is that the study only compared the percentage of different immune cells corresponding to various gene alterations. It is unclear how and why the immune cells are regulated by those genes.
Conclusions
It remains to be further investigated how these neuronal development genes and other axon guidance genes coordinate to modulate the TME. Different neuronal development genes may regulate the trafficking of various immune cells. Similar to axon guidance, these genes may guide the trafficking of immune cells. Tumor cells have acquired the ectopic expression of neuronal development genes and axon guidance genes; therefore, they have acquired the function of axon guidance genes ectopically. Similar to axon guidance molecules (which can function as nerve repellants or attractants depending on the subtype), genetic alterations in neuronal development genes in tumor cells allow the tumor cells to repel specific immune cells while attracting other immune cells. Such a hypothesis would need further testing with functional assays to measure the repellent and attractant function against tumors.
Acknowledgments
Funding: This work was supported in part by NIH grant R01 CA169702, NIH grant R01 CA197296, and Sidney Kimmel Comprehensive Cancer Center Support Grant P30 CA006973.
Footnote
Data Sharing Statement: Available at https://apc.amegroups.com/article/view/10.21037/apc-23-13/dss
Peer Review File: Available at https://apc.amegroups.com/article/view/10.21037/apc-23-13/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://apc.amegroups.com/article/view/10.21037/apc-23-13/coif). L.Z. was supported by NIH grant R01 CA169702, NIH grant R01 CA197296, and Sidney Kimmel Comprehensive Cancer Center Support Grant P30 CA006973. L.Z. serves as the Editor-in-Chief of Annals of Pancreatic Cancer. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of Johns Hopkins Medicine (J1568). These specimens were archived from two clinical trials (NCT02451982; NCT02648282), and written informed consent to allow the usage of archived specimens for other studies (the Johns Hopkins Medical Institution Institutional Review Board approved protocols at the Johns Hopkins Pancreatic Cancer Precision Medicine Center of Excellence Program) including this study was obtained from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rahib L, Wehner MR, Matrisian LM, et al. Estimated Projection of US Cancer Incidence and Death to 2040. JAMA Netw Open 2021;4:e214708. [Crossref] [PubMed]
- Muth ST, Saung MT, Blair AB, et al. CD137 agonist-based combination immunotherapy enhances activated, effector memory T cells and prolongs survival in pancreatic adenocarcinoma. Cancer Lett 2021;499:99-108. [Crossref] [PubMed]
- Cai J, Chen H, Lu M, et al. Advances in the epidemiology of pancreatic cancer: Trends, risk factors, screening, and prognosis. Cancer Lett 2021;520:1-11. [Crossref] [PubMed]
- Gu Z, Du Y, Zhao X, et al. Tumor microenvironment and metabolic remodeling in gemcitabine-based chemoresistance of pancreatic cancer. Cancer Lett 2021; Epub ahead of print. [Crossref] [PubMed]
- Principe DR, Korc M, Kamath SD, et al. Trials and tribulations of pancreatic cancer immunotherapy. Cancer Lett 2021;504:1-14. [Crossref] [PubMed]
- Xu X, Zhao Z, Guo S, et al. Increased semaphorin 3c expression promotes tumor growth and metastasis in pancreatic ductal adenocarcinoma by activating the ERK1/2 signaling pathway. Cancer Lett 2017;397:12-22. [Crossref] [PubMed]
- Zhang JF, Tao LY, Yang MW, et al. CD74 promotes perineural invasion of cancer cells and mediates neuroplasticity via the AKT/EGR-1/GDNF axis in pancreatic ductal adenocarcinoma. Cancer Lett 2021;508:47-58. [Crossref] [PubMed]
- Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399-405. [Crossref] [PubMed]
- Klagsbrun M, Eichmann A. A role for axon guidance receptors and ligands in blood vessel development and tumor angiogenesis. Cytokine Growth Factor Rev 2005;16:535-48. [Crossref] [PubMed]
- Chédotal A, Kerjan G, Moreau-Fauvarque C. The brain within the tumor: new roles for axon guidance molecules in cancers. Cell Death Differ 2005;12:1044-56. [Crossref] [PubMed]
- Adams RH, Eichmann A. Axon guidance molecules in vascular patterning. Cold Spring Harb Perspect Biol 2010;2:a001875. [Crossref] [PubMed]
- Nakanishi Y, Kang S, Kumanogoh A. Neural guidance factors as hubs of immunometabolic cross-talk. Int Immunol 2021;33:749-54. [Crossref] [PubMed]
- Liu D, Li J, Qi F, et al. Semaphorins and their receptors in pancreatic cancer: Mechanisms and therapeutic opportunities. Front Oncol 2022;12:1106762. [Crossref] [PubMed]
- Zheng L, Foley K, Huang L, et al. Tyrosine 23 phosphorylation-dependent cell-surface localization of annexin A2 is required for invasion and metastases of pancreatic cancer. PLoS One 2011;6:e19390. [Crossref] [PubMed]
- Foley K, Rucki AA, Xiao Q, et al. Semaphorin 3D autocrine signaling mediates the metastatic role of annexin A2 in pancreatic cancer. Sci Signal 2015;8:ra77. [Crossref] [PubMed]
- Jurcak NR, Rucki AA, Muth S, et al. Axon Guidance Molecules Promote Perineural Invasion and Metastasis of Orthotopic Pancreatic Tumors in Mice. Gastroenterology 2019;157:838-850.e6. [Crossref] [PubMed]
- Hwang WL, Jagadeesh KA, Guo JA, et al. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nat Genet 2022;54:1178-91. [Crossref] [PubMed]
- Li K, Tandurella JA, Gai J, et al. Multi-omic analyses of changes in the tumor microenvironment of pancreatic adenocarcinoma following neoadjuvant treatment with anti-PD-1 therapy. Cancer Cell 2022;40:1374-1391.e7. [Crossref] [PubMed]
- Yoo T, Kim SG, Yang SH, et al. A DLG2 deficiency in mice leads to reduced sociability and increased repetitive behavior accompanied by aberrant synaptic transmission in the dorsal striatum. Mol Autism 2020;11:19. [Crossref] [PubMed]
- Ali S, Hoven A, Dress RJ, et al. Identification of a novel Dlg2 isoform differentially expressed in IFNβ-producing plasmacytoid dendritic cells. BMC Genomics 2018;19:194. [Crossref] [PubMed]
- Keane S, Herring M, Rolny P, et al. Inflammation suppresses DLG2 expression decreasing inflammasome formation. J Cancer Res Clin Oncol 2022;148:2295-311. [Crossref] [PubMed]
- Górka B, Skubis-Zegadło J, Mikula M, et al. NrCAM, a neuronal system cell-adhesion molecule, is induced in papillary thyroid carcinomas. Br J Cancer 2007;97:531-8. [Crossref] [PubMed]
- Sakurai T. The role of NrCAM in neural development and disorders--beyond a simple glue in the brain. Mol Cell Neurosci 2012;49:351-63. [Crossref] [PubMed]
- Huang FJ, Liu YL, Wang J, et al. LncRNA RUNX1-IT1 affects the differentiation of Th1 cells by regulating NrCAM transcription in Graves' disease. Cell Cycle 2022;21:921-33. [Crossref] [PubMed]
- Yuan H, Wang Q, Liu Y, et al. A rare exonic NRXN3 deletion segregating with neurodevelopmental and neuropsychiatric conditions in a three-generation Chinese family. Am J Med Genet B Neuropsychiatr Genet 2018;177:589-95. [Crossref] [PubMed]
- Zheng JJ, Li WX, Liu JQ, et al. Low expression of aging-related NRXN3 is associated with Alzheimer disease: A systematic review and meta-analysis. Medicine (Baltimore) 2018;97:e11343. [Crossref] [PubMed]
- Al Shweiki MR, Oeckl P, Steinacker P, et al. Proteomic analysis reveals a biosignature of decreased synaptic protein in cerebrospinal fluid of major depressive disorder. Transl Psychiatry 2020;10:144. [Crossref] [PubMed]
- Nie S, Ni N, Chen N, et al. Development of a necroptosis-related gene signature and the immune landscape in ovarian cancer. J Ovarian Res 2023;16:82. [Crossref] [PubMed]
- Wang Z, Liu W, Chen C, et al. Low mutation and neoantigen burden and fewer effector tumor infiltrating lymphocytes correlate with breast cancer metastasization to lymph nodes. Sci Rep 2019;9:253. [Crossref] [PubMed]
- Caligiuri MA. Human natural killer cells. Blood 2008;112:461-9. [Crossref] [PubMed]
- Raskov H, Orhan A, Salanti A, et al. Natural Killer Cells in Cancer and Cancer Immunotherapy. Cancer Lett 2021;520:233-42. [Crossref] [PubMed]
- Sasca D, Szybinski J, Schüler A, et al. NCAM1 (CD56) promotes leukemogenesis and confers drug resistance in AML. Blood 2019;133:2305-19. [Crossref] [PubMed]
- Bartus K, Galino J, James ND, et al. Neuregulin-1 controls an endogenous repair mechanism after spinal cord injury. Brain 2016;139:1394-416. [Crossref] [PubMed]
- Ogier C, Colombo PE, Bousquet C, et al. Targeting the NRG1/HER3 pathway in tumor cells and cancer-associated fibroblasts with an anti-neuregulin 1 antibody inhibits tumor growth in pre-clinical models of pancreatic cancer. Cancer Lett 2018;432:227-36. [Crossref] [PubMed]
- Jonna S, Feldman RA, Swensen J, et al. Detection of NRG1 Gene Fusions in Solid Tumors. Clin Cancer Res 2019;25:4966-72. [Crossref] [PubMed]
- Zhu Y, Zhang H, Han X, et al. STAT3 mediated upregulation of C-MET signaling acts as a compensatory survival mechanism upon EGFR family inhibition in chemoresistant breast cancer cells. Cancer Lett 2021;519:328-42. [Crossref] [PubMed]
- Alizadeh A, Santhosh KT, Kataria H, et al. Neuregulin-1 elicits a regulatory immune response following traumatic spinal cord injury. J Neuroinflammation 2018;15:53. [Crossref] [PubMed]
- Bian-Fang Y, Dong-Ning W, Dan T, et al. The Role of Autophagy in Tumor Immune Infiltration in Colorectal Cancer. Anal Cell Pathol (Amst) 2022;2022:2055676. [Crossref] [PubMed]
- Zhao XT, Zhu Y, Zhou JF, et al. Development of a novel 7 immune-related genes prognostic model for oral cancer: A study based on TCGA database. Oral Oncol 2021;112:105088. [Crossref] [PubMed]
- Yuan D, Huang J, Yuan X, et al. Zinc finger protein 667 expression is upregulated by cerebral ischemic preconditioning and protects cells from oxidative stress. Biomed Rep 2013;1:534-8. [Crossref] [PubMed]
- Han D, Zhang C, Fan WJ, et al. Myocardial ischemic preconditioning upregulated protein 1(Mipu1):zinc finger protein 667 - a multifunctional KRAB/C2H2 zinc finger protein. Braz J Med Biol Res 2015;48:1-5. [Crossref] [PubMed]
- Li YZ, Chao R, Qu SL, et al. ZNF667 Suppressed LPS-induced Macrophages Inflammation through mTOR-dependent Aerobic Glycolysis Regulation. Curr Pharm Des 2023;29:1361-9. [Crossref] [PubMed]
- Bohuslavova R, Cerychova R, Papousek F, et al. HIF-1α is required for development of the sympathetic nervous system. Proc Natl Acad Sci U S A 2019;116:13414-23. [Crossref] [PubMed]
- Paredes F, Williams HC, San Martin A. Metabolic adaptation in hypoxia and cancer. Cancer Lett 2021;502:133-42. [Crossref] [PubMed]
- Wang X, Zhao D, Xie H, et al. Interplay of long non-coding RNAs and HIF-1α: A new dimension to understanding hypoxia-regulated tumor growth and metastasis. Cancer Lett 2021;499:49-59. [Crossref] [PubMed]
- Kajioka H, Kagawa S, Ito A, et al. Targeting neutrophil extracellular traps with thrombomodulin prevents pancreatic cancer metastasis. Cancer Lett 2021;497:1-13. [Crossref] [PubMed]
- McGettrick AF, O'Neill LAJ. The Role of HIF in Immunity and Inflammation. Cell Metab 2020;32:524-36. [Crossref] [PubMed]
- Li HH, Liu Y, Chen HS, et al. PDGF-BB-Dependent Neurogenesis Buffers Depressive-Like Behaviors by Inhibition of GABAergic Projection from Medial Septum to Dentate Gyrus. Adv Sci (Weinh) 2023;10:e2301110. [Crossref] [PubMed]
- Yang Y, Mao W, Wang L, et al. Circular RNA circLMF1 regulates PDGF-BB-induced proliferation and migration of human aortic smooth muscle cells by regulating the miR-125a-3p/VEGFA or FGF1 axis. Clin Hemorheol Microcirc 2022;80:167-83. [Crossref] [PubMed]
- Fernandez RF, Ellis JM. Acyl-CoA synthetases as regulators of brain phospholipid acyl-chain diversity. Prostaglandins Leukot Essent Fatty Acids 2020;161:102175. [Crossref] [PubMed]
- Zhu Z, Wang D, Shen Y. Loss of ACSM3 confers worsened prognosis and immune exclusion to cutaneous melanoma. J Cancer 2020;11:6582-90. [Crossref] [PubMed]
- Dinamarca MC, Raveh A, Schneider A, et al. Complex formation of APP with GABA(B) receptors links axonal trafficking to amyloidogenic processing. Nat Commun 2019;10:1331. [Crossref] [PubMed]
- Ge L, Hoa NT, Wilson Z, et al. Big Potassium (BK) ion channels in biology, disease and possible targets for cancer immunotherapy. Int Immunopharmacol 2014;22:427-43. [Crossref] [PubMed]
- Bailey CS, Moldenhauer HJ, Park SM, et al. KCNMA1-linked channelopathy. J Gen Physiol 2019;151:1173-89. [Crossref] [PubMed]
- Shek D, Chen D, Read SA, et al. Examining the gut-liver axis in liver cancer using organoid models. Cancer Lett 2021;510:48-58. [Crossref] [PubMed]
- Goggolidou P, Richards T. The genetics of Autosomal Recessive Polycystic Kidney Disease (ARPKD). Biochim Biophys Acta Mol Basis Dis 2022;1868:166348. [Crossref] [PubMed]
- Zhang MZ, Mai W, Li C, et al. PKHD1 protein encoded by the gene for autosomal recessive polycystic kidney disease associates with basal bodies and primary cilia in renal epithelial cells. Proc Natl Acad Sci U S A 2004;101:2311-6. [Crossref] [PubMed]
- Reznikov LR. Cystic Fibrosis and the Nervous System. Chest 2017;151:1147-55. [Crossref] [PubMed]
- Stevens B, Allen NJ, Vazquez LE, et al. The classical complement cascade mediates CNS synapse elimination. Cell 2007;131:1164-78. [Crossref] [PubMed]
- Schafer DP, Lehrman EK, Kautzman AG, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 2012;74:691-705. [Crossref] [PubMed]
- Bialas AR, Stevens B. TGF-β signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat Neurosci 2013;16:1773-82. [Crossref] [PubMed]
- Sekar A, Bialas AR, de Rivera H, et al. Schizophrenia risk from complex variation of complement component 4. Nature 2016;530:177-83. [Crossref] [PubMed]
- Chen YZ, Matsushita MM, Robertson P, et al. Autosomal dominant familial dyskinesia and facial myokymia: single exome sequencing identifies a mutation in adenylyl cyclase 5. Arch Neurol 2012;69:630-5. [Crossref] [PubMed]
- Pieroni JP, Miller D, Premont RT, et al. Type 5 adenylyl cyclase distribution. Nature 1993;363:679-80. [Crossref] [PubMed]
- Ferrini A, Steel D, Barwick K, et al. An Update on the Phenotype, Genotype and Neurobiology of ADCY5-Related Disease. Mov Disord 2021;36:1104-14. [Crossref] [PubMed]
- Cummins TR, Dib-Hajj SD, Waxman SG. Electrophysiological properties of mutant Nav1.7 sodium channels in a painful inherited neuropathy. J Neurosci 2004;24:8232-6. [Crossref] [PubMed]
- Black JA, Frézel N, Dib-Hajj SD, et al. Expression of Nav1.7 in DRG neurons extends from peripheral terminals in the skin to central preterminal branches and terminals in the dorsal horn. Mol Pain 2012;8:82. [Crossref] [PubMed]
- Sui Q, Peng J, Han K, et al. Voltage-gated sodium channel Na(v)1.5 promotes tumor progression and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cancer Lett 2021;500:119-31. [Crossref] [PubMed]
- Dib-Hajj SD, Black JA, Waxman SG. Voltage-gated sodium channels: therapeutic targets for pain. Pain Med 2009;10:1260-9. [Crossref] [PubMed]
- Waxman SG. Neuroscience: Channelopathies have many faces. Nature 2011;472:173-4. [Crossref] [PubMed]
- Lonic A, Powell JA, Kong Y, et al. Phosphorylation of serine 779 in fibroblast growth factor receptor 1 and 2 by protein kinase C(epsilon) regulates Ras/mitogen-activated protein kinase signaling and neuronal differentiation. J Biol Chem 2013;288:14874-85. [Crossref] [PubMed]
- Barbagallo D, Palermo CI, Barbagallo C, et al. Competing endogenous RNA network mediated by circ_3205 in SARS-CoV-2 infected cells. Cell Mol Life Sci 2022;79:75. [Crossref] [PubMed]
- Li X, Ding W, Rao Y, et al. Role of protein tyrosine phosphatase receptor type M in epithelial ovarian cancer progression. J Ovarian Res 2023;16:131. [Crossref] [PubMed]
- Brady-Kalnay SM, Mourton T, Nixon JP, et al. Dynamic interaction of PTPmu with multiple cadherins in vivo. J Cell Biol 1998;141:287-96. [Crossref] [PubMed]
- Burden-Gulley SM, Brady-Kalnay SM. PTPmu regulates N-cadherin-dependent neurite outgrowth. J Cell Biol 1999;144:1323-36. [Crossref] [PubMed]
- Nair R, Lauks J, Jung S, et al. Neurobeachin regulates neurotransmitter receptor trafficking to synapses. J Cell Biol 2013;200:61-80. [Crossref] [PubMed]
- Gromova KV, Muhia M, Rothammer N, et al. Neurobeachin and the Kinesin KIF21B Are Critical for Endocytic Recycling of NMDA Receptors and Regulate Social Behavior. Cell Rep 2018;23:2705-17. [Crossref] [PubMed]
- Medrihan L, Rohlmann A, Fairless R, et al. Neurobeachin, a protein implicated in membrane protein traffic and autism, is required for the formation and functioning of central synapses. J Physiol 2009;587:5095-106. [Crossref] [PubMed]
Cite this article as: Mu K, Fu J, Gai J, Ravichandran H, Zheng L, Sun WC. Genetic alterations in the neuronal development genes are associated with changes of the tumor immune microenvironment in pancreatic cancer. Ann Pancreat Cancer 2023;6:10.


